 For anyone who uses python and ds9 to visualize their FITS files, I think pyds9 is now a must-have. It is officially written and developed through SAOImage ds9 so it will be supported for the long haul. Here are the primary links to get going:
For anyone who uses python and ds9 to visualize their FITS files, I think pyds9 is now a must-have. It is officially written and developed through SAOImage ds9 so it will be supported for the long haul. Here are the primary links to get going:
The documentation is pretty basic so I put together a very small tutorial of a way I might typically use it. Most of the controls work through the XPA interface, which seems very flexible, if a little tricky to figure out exactly how to make it work for specific use-cases. If you have samples of how you use pyds9 or ds9 xpa, post them in the comments or on this wiki page.
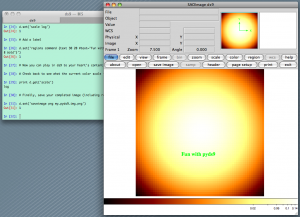
from scipy import stats import numpy as np import ds9 # Make a 2D gaussian image that is stored in a 2D numpy array x = np.arange(-3, 3, 0.1) xx, yy = np.meshgrid(x, x) gauss2d = stats.norm.pdf(xx) * stats.norm.pdf(yy) # Now open ds9 (this assumes no ds9 instance is yet running) d = ds9.ds9() # Load up our 2D gaussian d.set_np2arr(gauss2d) # Zoom to fit d.set('zoom to fit') # Change the colormap and scaling d.set('cmap bb') d.set('scale log') # Add a label d.set('regions command {text 30 20 #text="Fun with pyds9" font="times 18 bold"}') # Now you can play in ds9 to your heart's content. # Check back to see what the current color scale is. print d.get('scale') # Finally, save your completed image (including regions or labels) d.set('saveimage png my_pyds9_img.png') |
The great thing about pyds9 is that it makes it easy to write your own quick analysis tasks – I’m sure there must be others who have been desperate for simple aperture photometry in ds9. My attempt at that is http://code.google.com/p/agpy/source/browse/trunk/agpy/ds9_phot.py plus http://code.google.com/p/agpy/source/browse/trunk/agpy/phot.ds9.ans.
I have found XPA useful without python for making DS9 figures reproducible. DS9 is capable of making publication-quality figures, but it requires a lot of button clicks… XPA allows you to script it.
Jessica, I’ll take you as an expert and ask you: Have you found a way to plot points, not boxes, actual solid points one pixel-wide? If you have worked with the IRAF, I’m interested in reproducing the behaviour of tvmark to visually identify bad pixels, for example.
I’ve asked Jessica, but if anybody has the answer, please share.
Thanks!
I wrote a couple of routines that use pyds9 to read image data from regions (http://bitbucket.org/nhmc/pyserpens/src/tip/ds9utils.py).
It’s very cool to be able to load an image inside ds9, then use the python interpreter to play around with the data inside regions you define using ds9.
I’m not sure how to plot a one-pixel wide solid point, but regions can just be a single cross or plus sign (try them out in the region – shape menu).
Neil – for region parsing, there’s a pretty complete package called “pyregion” available from http://github.com/leejjoon/pyregion and/or http://leejjoon.github.com/pyregion/ (while the URLs look quite similar, they take you to very different front-pages).
Kathleen – The best I can think of is replacing the bad data in the array with NAN and re-displaying it. You can set the NAN color to be anything in the Edit:Preferences menu.
In principle you should be able to use tvmark as well:
In practice, I haven’t gotten it to work, but then again, I have never used tvmark before.
Thanks for the suggestions. I see that it isn’t something I missed, it is just not implemented.
Have you ever used numdisplay?
http://stsdas.stsci.edu/numdisplay/
It has been around a long time and is well supported by STScI. It works well with Pyraf.
Yes, Chris, I’ve used numdisplay extensively. It does exactly what I need it to do. The only problem was that it was too slow to draw hundreds of points. I think you guys improved the performance following the report I made. I haven’t had the chance to test it out again.
Is anyone else unable to manipulate the scale with the mouse in the normal ds9 way when using pyds9?
I keep getting this message every time I run ds9().
” Traceback (most recent call last):
File “test.py”, line 11, in
d = ds9.ds9()
File “/opt/local/Library/Frameworks/Python.framework/Versions/2.7/lib/python2.7/site-packages/ds9.py”, line 269, in __init__
raise ValueError, ‘no active ds9 running for target: %s’ % target
ValueError: no active ds9 running for target: ds9″
Am I doing something wrong here??
Thanks!
In Preference menu of ds9, ‘Initialize XPA’ needs to be ticked to use the xpa messaging system. This was not ticked in my ds9 for some reason.. Now it works fine!
Cheers,
I am also getting the same error. I have the ‘initialize XPA’ already turned on. And still I am getting this error. Can I get some help on this..
Thanks
Hi all, I’ve created a lightweight wrapper to pyds9, which adds some ready made python wrappers to common actions such as pan, zoom, scale, etc. I also use a bit of python trickery so you can write these functions for a single frame, then automatically apply them to all frames if you so desire. It’s a work in progress rather than a polished product, but it should be easy for anyone with a little python / ds9 knowledge to get stuck in. Collaborators welcome!
https://github.com/timstaley/pyds9_ex
Hi, Tim, can you give me a tip?
I want to save an image with custom resolution and keeping drawed regions.
Thanks,
Manuel Olmedo
Hi,
Can anybody help me to figure out what I do wrong in my function? I just want to open DS9 and display the 4th extension of the FITS file like:
def openFFI(infile):
hdulist = pyfits.open(infile)
process_DS9 = subprocess.Popen(‘ds9’, shell=True)
time.sleep(5)
# I initialize the last opened DS9 here
ds9_lastInstance = ds9_targets()[len(ds9_targets())-1].split(‘ ‘)[1]
d = DS9(ds9_lastInstance)
d.set_pyfits(hdulist[4])
This is the output error message:
————————————-
Traceback (most recent call last):
File “”, line 1, in
File “my_kepffi.py”, line 57, in openFFI
d.set_pyfits(hdulist[4])
File “/usr/local/lib/python2.7/site-packages/pyds9.py”, line 471, in set_pyfits
raise ValueError, ‘requires astropy.HDUList as input’
ValueError: requires astropy.HDUList as input
————————————-
thanks in advance
oliver